GRCh38.p14 (GCA_000001405.29/GCF_000001405.40), the latest update to the human reference assembly, has been released! It adds 69 new patch scaffolds, 51 of which are FIX patches that update sequences on the GRCh38 reference chromosomes or alternate loci, while 18 are NOVEL patches, providing new alternate representations for complex genomic regions that are inadequately represented by a single sequence. Two previously released FIX patches were also updated. With this release, the reference assembly contains a total of 250 patch scaffolds (164 FIX, 90 NOVEL).
30 of the patches included in this release include genome updates made in support of the MANE project, a joint NCBI-EBI effort to produce a minimal set of matching RefSeq and Ensembl transcripts of protein coding genes, creating a matched pair of transcripts but retaining their respective identifiers. Read more about MANE effort in their recent
Nature publication. The corresponding patch updates to the reference assembly involved changes addressing normal human variation as well as correcting errors in the underlying component sequences.
Of the 53 FIX patches in GRCh38.p14, 23 of these correct errors in individual assembly component sequences, resulting in updates to 12 gene representations (Table 1). 20 are variation-related updates, 12 of which provide the coding allele for 13 polymorphic pseudogenes that are non-coding on the corresponding GRCh38 chromosomes (Table 2). Additionally, 2 provided sequence updates at chromosomal loci where it's unclear if the GRCh38 sequence is in error or a rare haplotype. Patch scaffolds in GRCh38.p14 close 6 gaps in the reference assembly, and extend sequence into one other gap. 4 of the closed gaps are located within chromosomes, while the remaining 2 patch scaffolds closed "pre-telomeric" gaps, extending the sequence of the chromosome into the telomeric repeats.
 |
Table 1. Gene representations updated on FIX patches addressing assembly component problems.
|
 |
| Table 2. Coding alleles of polymorphic pseudogenes updated by FIX patches addressing genomic variation. |
An example of an important FIX patch in this release is an update to APOB, one of the genes the American College of Medical Genetics and Genomics recommends for reporting of incidental findings in clinical exome and genome sequencing. The patch scaffold provided in GRCh38.p14 represents the common allele.
There are 18 NOVEL patches in this release, providing alternate sequence representations of chromosomal sequences, including 9 genes (Table 3). Other NOVEL patches represent inversion and insertion haplotypes relative to the corresponding chromosomal region.
 |
| Table 3. Genes with alternate sequence representation on GRCh38.p14 NOVEL patches. |
Shown below is an example of an update to
PRDM9, a medically important gene in which naturally occuring allelic variation regulates the activity of meiotic recombination hotspots. The original GRCh38 release represents the relatively rare "B" allele on chromosome 5. With the release of GRCh38.p14, a NOVEL patch scaffold has been added to the assembly (
MU273356.1/
NW_025791779.1) to provide additional representation for the sequence of the more common "A" allele.
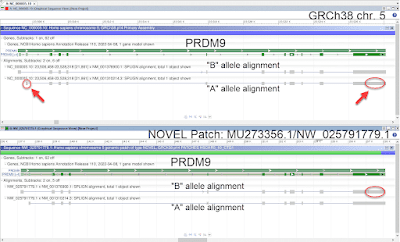 |
Figure 1. PRDM9 allele representation in GRCh38.p14. Top: Alignment of PRDM9 "A" (NM_001310214.3) and "B" (NM_001376900.1) allele transcripts to chromosome 5. The chromosome sequence represents the "B" allele. The red circles and arrows highlight mismatches in the alignment of the "A" allele. Bottom: Alignment of Alignment of PRDM9 "A" and "B" allele transcripts to the NOVEL patch added in GRCh38.p14. The patch represents the "A" allele. The red circle highlights mismatches in the alignment of the "B" allele.
|
Notably, 9 of the NOVEL patches used clone sequence generated by Evan Eichler's lab as part of a published study of the evolution and population diversity of human-specific segmental duplications.The GRC also used sequences generated by the Eichler lab to create a FIX patch to improve a GRCh38 chromosome 5 alternate locus scaffold (KI270897.1/NT_187651.1) representing the haplotype from the CHM1 hydatidiform mole at the hypervariable SMA locus. Informed by CHM1 Bionano optical map data, the GRC provided a FIX patch (MU273354.1/NW_025791777.1) that corrects component order and adds sequence from several newly sequenced CHM1 BAC clones to the alternate locus scaffold.
 |
| Figure 2. A FIX patch corrects the sequence path of the GRCh38 alt locus scaffold providing representation of the CHM1 haplotype for the SMA region on chromosome 5. Top: Tiling path of component clones in the alt loci scaffold. Middle: Tiling path of component clones in the FIX patch scaffold. Blue outline: clones excluded from fix scaffold. Green outline: clones added to fix scaffold. Magenta outline: clones from alt scaffold retained in fix scaffold. Black: sequence gap. Bottom: Alignment of fix patch scaffold path to CHM1 Bionano optical map, demonstrating concordance. |
This patch release also extends GRC efforts to identify and exclude problematic sequences, such as false redundancies and contamination, from the reference assembly. The companion BED file available from GenBank that identifies such regions and can be used as a mask to exclude them from analyses, has now been updated. The latest updates reflect curation done in response to reports from GRCh38 analyses performed by the Genome In a Bottle (GIAB) and Telomere-to-Telomere (T2T) consortia. In addition to the chromosome 21p regions previously reported, the file provides coordinates for 7 other regions in which the sequence falsely duplicates other sequence found in the assembly.
We are grateful to our community collaborators for the sequences and analyses that contributed to the updates in GRCh38.p14. Please alert the GRC if you have specific assembly issues to report, or contact us for any questions or feedback. We'd love to hear from you!


