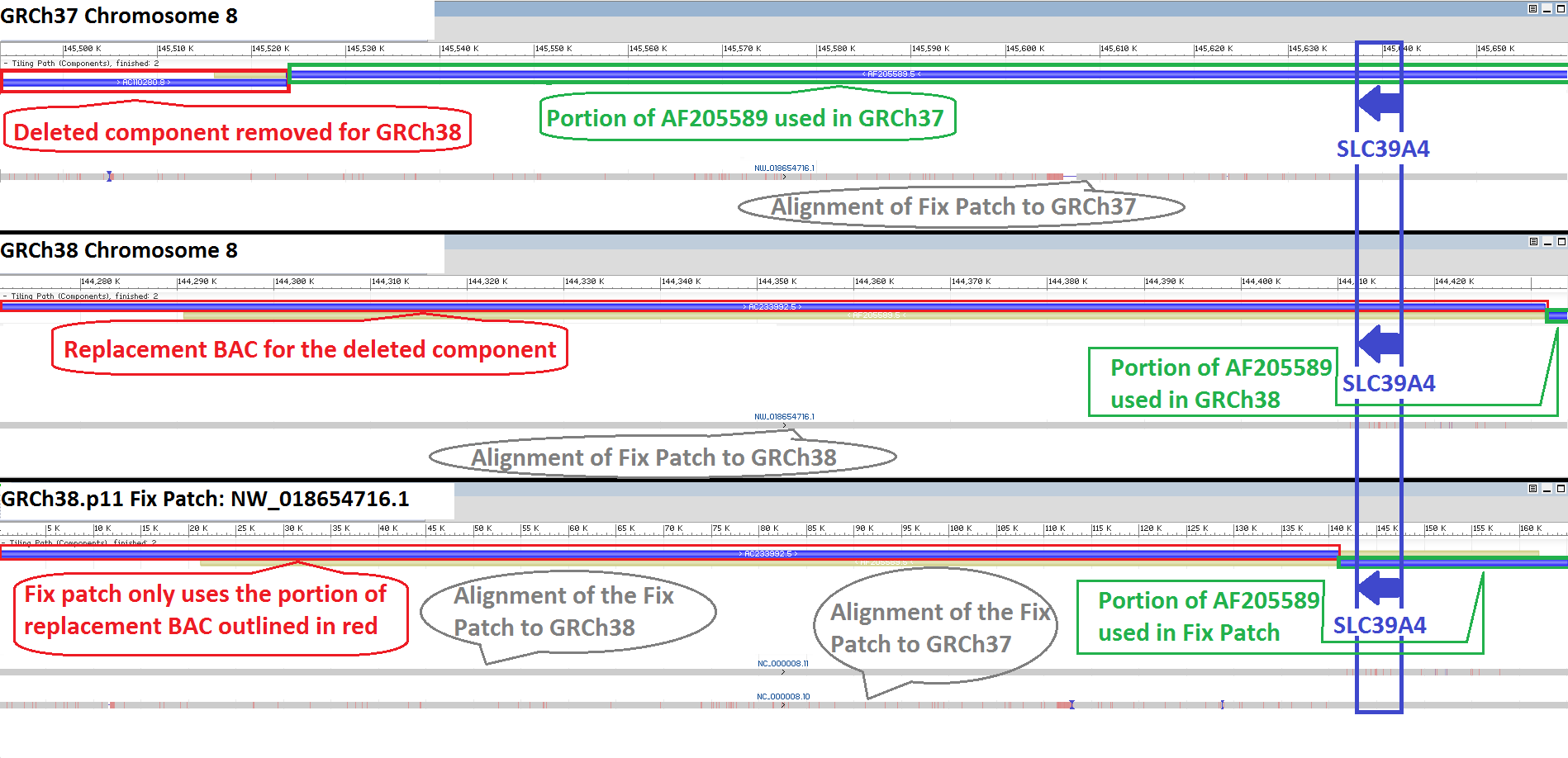
The GRCh38.p11 patch release includes the fix patch scaffold KZ208914.1/NW_018654716.1 that updates two bases in SLC39A4, a change prompted by user request and that has implications for clinically-relevant variant analyses of this gene. This GRCh38 fix patch restores the gene to the same representation found in GRCh37.
SLC39A4, solute carrier family 39 member 4 (NCBI Gene ID: 55630), encodes a protein that is required for dietary zinc absorption in the intestine. In GRCh37, the BAC clone AF205589.5 (CTA-393G12) provided the sequence for SLC39A4 on chromosome 8. In GRCh38, AC233992.5 (RP11-735F20) replaced AC110280.8 (CTD-3232M19), the BAC clone upstream and adjacent to AF205589.5. As a consequence of this change, the default switch point in the overlap between these two clones led to the newly added RP11 clone providing the underlying sequence for SLC39A4 in GRCh38. A user subsequently contacted the GRC to report that this GRCh38 update introduced changes at two bases, a G to C substitution at CM000670.2/NC_000008.11: 144,414,297 (rs1871534) and a G to A substitution at CM000670.2/NC_000008.11: 144,415,811 (rs1871533). The “A” allele in GRCh38 has a global MAF of only 0.0098, according to data from the 1000 Genomes project, making it a rare allele. Additionally, the GRCh38 haplotype confounds clinical prediction algorithms at this locus because the p.Leu372Val (rs1871534) allele found in GRCh38 is incompatible with the p.Leu372Pro variant, which is implicated in Acrodermatitis entropathica (PMID: 12032886).
Review of Illumina genome sequencing data from an RP11 paired end WGS library (SRR834589) aligned to GRCh38 confirms the sequence of AC233992.5, the RP11 BAC clone providing the SLC39A4 sequence in GRCh38. Even though there are no sequence errors at these locations, the GRC agreed to further update these sites to address the needs of clinical researchers and to reduce the presence of globally rare alleles in the reference. Changing the switch point between the two GRCh38 BAC components allows the clone AF205589.5 to once again provide the sequence for SLC39A4 (Figure 1), and review by RefSeq annotators confirmed that this update addresses the two bases without introducing other changes that negatively affect gene representation. These updated bases are now available in the fix patch, and will be incorporated into the primary chromosome at the next major assembly release (GRCh39, currently unscheduled).